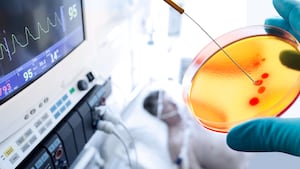
Superfund sites take many forms: abandoned mines, shuttered manufacturing facilities, former military bases, and, in the case of New York City’s infamous Gowanus Canal, waterways. These places are so contaminated with hazardous waste they pose a serious risk to human health and the Environmental Protection Agency has made cleaning them up a national priority.
Brooklyn’s Gowanus Canal has been a basin of industry waste for decades—arsenic from tanneries and runoff from cement and oil lamp factories. It’s still on the receiving end of Brooklyn’s sewage system and since 2014, at least three human corpses have been found in its waters. Scientists, however, think the Gowanus Canal and places like it could harbor new medicines.
Chris Mason, biochemist and chief researcher at Cornell University’s Mason Laboratory, is among them. He says the organisms that thrive among the pollution could hold treatments for any number of maladies, from cancer to AIDS. Faced with the rising threat of antibiotic resistance, Mason’s team is focused on sourcing new antibiotics from the sludge. So far, they believe they are close to isolating four new strains of antibiotics from compounds produced by the microbes living in the Gowanus Canal.
“It’s completely toxic but we knew things are living in there,” says Mason. These ‘things’ are called extremophiles, and they’re incredibly important to the future of medicine.
Extremophile is the name given to the kinds of (usually microscopic) creatures that thrive in Earth’s most hostile environments, places that would be a death sentence for most other life forms—deep sea vents, the Arctic, and Superfund sites.
In experiments, extremophiles have attacked invasive cells like cancer or harmful bacteria without harming healthy cells. That’s one of the reasons why scientists think the organisms that thrive in Superfund sites could lead to medical breakthroughs.
In 2015, Mason started working with the Brooklyn-based incubator BK Bioreactor to map the DNA of the microscopic creatures that thrive in the Gowanus Canal. Because the Superfund site has a rich history of polluters, the incubator wanted to sequence—a technology that reads and records the DNA of microscopic organisms—the canal’s tailor-made extremophiles in an effort to find better ways to clean it up. Mason thought the DNA samples could serve double-duty and began mining them for strings of DNA that could be medically useful; and he’s not the first to suspect a Superfund site might hold the key to tomorrow’s medical breakthroughs.
When research chemists Andrea and Don Stierle accepted positions at Montana State University in the early 1990s, they found themselves in a small mining town, drained of project funding, and less than a mile from the largest Superfund site in North America: the Berkeley Pit.
“You’re most creative at rock bottom,” says Andrea Stierle. “Every time we run out of funding is when we find something new because we don’t have anything to lose.”
The Stierles were the first to look for medicine in a Superfund site. They’ve isolated compounds from the Berkeley Pit that could lead to new treatments for lung cancer, ovarian cancer and leukemia.
Like Mason, the Stierles have directed their latest research to antibiotics.
For the first time, the husband-wife team is patenting a potential drug that was born out of their Superfund research. The potential antibiotic is created through a careful process that involves growing two types of fungi extracted from the Berkeley Pit. The marriage produces what the Stierles hope will be an answer to antibiotic-resistant infections.
So far, the compounds have successfully killed four strains of Methicillin-resistant Staphylococcus aureus—or MRSA—in a lab. Lab experiments are the lowest rung on the scientific experiment ladder (followed by animal studies and then studies done on humans) but it’s an important milestone since scientists have only just begun studying the life in Superfund sites.
According to the MRSA Research Center at the University of Chicago, around 20,000 fatal MRSA infections are reported in the U.S. each year. The skin infection was identified as the first case of antibiotic resistance and the infection remains the poster child of antibiotic resistance in the developed world today.
Though MRSA is actually on the decline in the U.S., a 2015 report by economist Jim O’Neill estimates annual deaths attributed to antibiotic resistance worldwide will climb to 10 million by 2050 if we don’t find new sources of antibiotics. A recent report from the United Nations echoes that prediction.
Mason believes the world is already in dire trouble when it comes to antibiotic resistance, but says there’s a silver lining. He’s hopeful the new age of data will hold the key to reversing the threat of antibiotic resistance.
Mason’s team compares the newly sequenced DNA of extremophiles to that of known antibiotics. If they find a sequence of DNA that looks like a match, the next step is to grow that organism and see how it reacts with bacteria.
Other scientists have done the same with microorganisms living in other extreme places. Anna-Louise Reysenbach, an extremophile biologist at the University of Portland, and her team sequenced the DNA of microscopic organisms living in deep-sea heat vents. Their research focused on better understanding archaea.
Archaea are often classified as extremophiles. They live in places that are too hot, too dark, or too toxic for any other life to survive, like in heat vents on the ocean floor or in Superfund sites.
“They don’t just survive,” says Reysenbach. “They’re happy, they thrive.”
Archaea are one type of Gowanus Canal life (bacteria and fungi are also included) that Mason’s team analyzes. They’re important to the future of medicine mostly because we just don’t know a lot about them.
Archaea weren’t recognized by the scientific community until the late 1970s. Before that, scientists thought there were only two types of life on Earth: procaryotes, organisms that do not have a nucleus, mostly bacteria; and eucaryotes, anything with a nucleus, basically all life that is familiar to us, including humans.
Archaea were originally thought to be procaryotes, until scientists realized that bacteria that thrived in high temperatures or produced methane were genetically distinct from other procaryotes, so they designated a third category—an entirely new domain of life that we still don’t know much about. And therein lies the promise: We’ve scoured the rainforest for medicine and even the mold that grows on our food, but we still don’t know much about archaea and other extremophiles. Places like deep sea vents and Superfund sites, and the organisms that live in them, have a lot of potential simply because scientists haven’t explored them yet.
New technology like Next-Generation Sequencing allows scientists to understand these places to an extent that wasn’t possible until now. The world is experiencing a renaissance of biological discovery.
“It could have been really bad,” says Mason, referencing the looming threat of antimicrobial resistance, “but we’ve never been so equipped to address the problem.”